Advanced spectral imaging for precise biological analysis
Lightsheet microscopy and confocal imaging of thick tissues generate complex 3D images that are often difficult or impossible to analyze using classical software such as ImageJ or QuPath.
To address this challenge, we leverage advanced tools and programming techniques to enable precise spatial analysis in three dimensions.
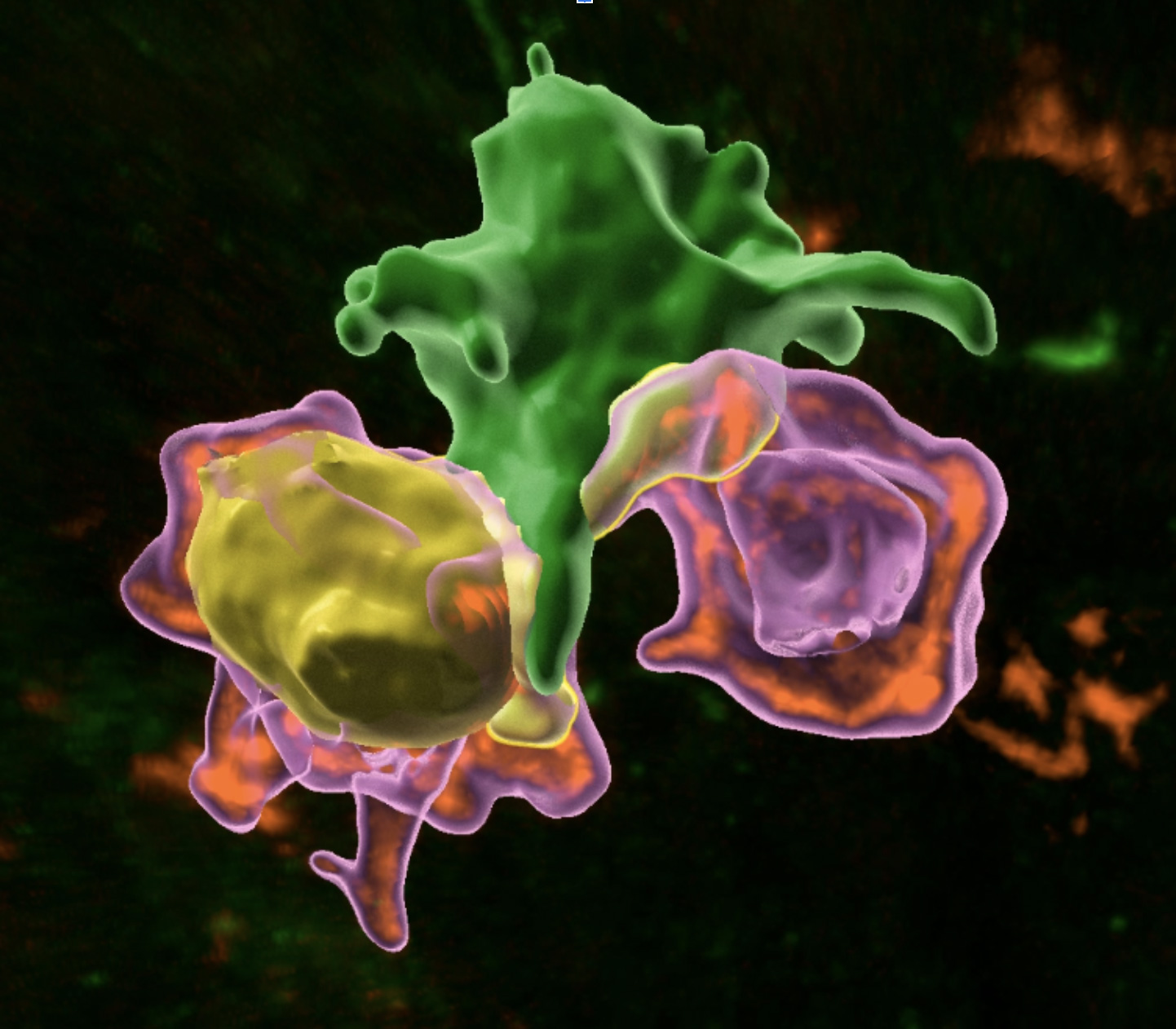
*A phagocyte (green) interacts with two T cells (magenta), one of which is proliferating (yellow) - Hugues Lelouard and Mathieu Fallet
Applications
The platform uses Imaris software (Oxford Instruments), the most widely used tool in biology for 3D spatial analysis, with three full licenses available on the PICsL node. For dedicated projects requiring specific solutions, we also implement Python programming, Napari (interactive image viewer), and Cellpose (cellular segmentation).
Available systems:
At CIML:
For example, at CIML, current projects focus on deciphering cell interactions in 3D, specifically analyzing the random contributions of interactions caused by cell density.
At IBDM:
8 high-performance PCs with GPU capabilities (8 GB, 32 GB, 48 GB) to support rapid finetuning and training of new neural network models for advanced 3D analysis.
The 3D image analysis platform at PICsL integrates cutting-edge software and hardware resources, enabling researchers to tackle the complexities of three-dimensional biological data with precision and efficiency.

Enables advanced 3D spatial analysis of complex biological images.

Utilizes Imaris software with three full licenses available.

Supports Python programming, Napari, and Cellpose for specific projects.

Eight high-performance GPUs enable neural network training and finetuning.









